Example 5: Using a real datasets (DES)
Fit halo mass to shear profile using DES data
the LSST-DESC CLMM team
This notebook can be run on NERSC.
Here we demonstrate how to run CLMM on real observational datasets. As an example, we use the data from the Dark Energy Survey (DES) public releases. The catalogs can be accessed from the NOIRLab Astro Data Lab.
The steps in this notebook includes: - Setting things up - Selecting a cluster - Downloading the published catalog at the cluster field - Loading the catalog into CLMM - Running CLMM on the dataset
Acknowledgement
DES data: https://des.ncsa.illinois.edu/thanks
Astro Data Lab: https://datalab.noirlab.edu/acknowledgements.php
## 1. Setup
We import packages.
import numpy as np
import matplotlib.pyplot as plt
from astropy.table import Table
import pickle as pkl
from pathlib import Path
## 2. Selecting a cluster
We use the DES Y1 redMaPPer Catalogs (https://des.ncsa.illinois.edu/releases/y1a1/key-catalogs/key-redmapper) to select a list of high-richness (LAMBDA) galaxy clusters, which likely have high masses.
Name |
RA (deg) |
DEC (deg) |
Z_LAMBDA |
LAMBDA |
|---|---|---|---|---|
RMJ025415.5 585710.7 |
43.564574 |
-58.95297 |
0.429804 |
234.50368 |
RMJ051637.4 543001.6 |
79.155704 |
-54.500456 |
0.30416065 |
195.06956 |
RMJ224851.8 443106.3 |
342.215897 |
-44.518403 |
0.3514858 |
178.83827 |
## 3. Downloading the catalog at the cluster field
We consider RMJ051637.4-543001.6 (ACO S520) as an example. We can access
the DES catalog from NOIRLab Data Lab
(https://datalab.noirlab.edu/query.php?name=des_dr1.shape_metacal_riz_unblind).
No registration is required. We make the query and download the catalogs
in “Query Interface”. We use coadd_objects_id to cross match the
shape catalog and photo-z catalog
(https://datalab.noirlab.edu/query.php?name=des_dr1.photo_z). Since the
cluster is at redshift about 0.3, a radius of 0.3 deg would be about a
radial distance of 5 Mpc. The final catalog includes shape info and
photo-z. Here is an example of the query SQL command. The query could
take a few minutes and the size of the catalog is about 1.4 MB (.csv).
SELECT P.mean_z,
C.ra, C.dec, C.e1, C.e2, C.r11, C.r12, C.r21, C.r22
FROM des_dr1.photo_z as P
INNER JOIN des_dr1.shape_metacal_riz_unblind as C
ON P.coadd_objects_id=C.coadd_objects_id
WHERE 't' = Q3C_RADIAL_QUERY(C.ra, C.dec,79.155704, -54.500456, 0.3)
AND P.minchi2<1
AND P.z_sigma<0.1
AND C.flags_select=0
## 4. Loading the catalog into CLMM
Once we have the catalog, we read in the catalog, make cuts on the catalog, and adjust column names to prepare for the analysis in CLMM.
%%time
# Assume the downloaded catalog is at this path:
filename = "ACOS520_DES.csv"
catalog = filename.replace(".csv", ".pkl")
if not Path(catalog).is_file():
data_0 = Table.read(filename, format="ascii.csv")
pkl.dump(data_0, open(catalog, "wb"))
else:
data_0 = pkl.load(open(catalog, "rb"))
CPU times: user 0 ns, sys: 3.52 ms, total: 3.52 ms
Wall time: 3.12 ms
print(data_0.colnames)
['mean_z', 'ra', 'dec', 'e1', 'e2', 'r11', 'r12', 'r21', 'r22']
Shear response
Shears in the DES data have been measured using the metacal method
and the catalog provides the shear response terms
(\(r11,r22, r12, r21\)) required to calibrate the shear values.
print(np.mean(data_0["r11"]), np.mean(data_0["r22"]))
print(np.mean(data_0["r12"]), np.mean(data_0["r21"]))
r_diag = np.mean([np.mean(data_0["r11"]), np.mean(data_0["r22"])])
r_off_diag = np.mean([np.mean(data_0["r12"]), np.mean(data_0["r21"])])
print(r_diag, r_off_diag, r_off_diag / r_diag)
0.7105854684953065 0.7070498820143473
-0.017056656529555746 0.008455599747631376
0.7088176752548269 -0.004300528390962185 -0.006067185598068083
The diagonal terms are close to each other. The off-diagonal terms are much smaller (<1%). We use the mean of the diagonal terms to reduce noise. We also skip the selection bias since it is typically at percent level.
# Adjust column names.
def adjust_column_names(catalog_in):
# We consider a map between new and old column names.
# Note we have considered shear calibration here.
column_name_map = {
"ra": "ra",
"dec": "dec",
"z": "mean_z",
"e1": "e1",
"e2": "e2",
}
catalog_out = Table()
for i in column_name_map:
catalog_out[i] = catalog_in[column_name_map[i]]
catalog_out["e1"] /= r_diag
catalog_out["e2"] /= r_diag
return catalog_out
obs_galaxies = adjust_column_names(data_0)
select = obs_galaxies["e1"] ** 2 + obs_galaxies["e2"] ** 2 <= 1.0
print(np.sum(~select))
obs_galaxies = obs_galaxies[select]
0
Basic visualization
def make_plots(catalog_in):
# Scatter plot
plt.figure()
plt.scatter(catalog_in["ra"], catalog_in["dec"], c=catalog_in["z"], s=1.0, alpha=1)
cb = plt.colorbar()
plt.xlabel("ra")
plt.ylabel("dec")
cb.ax.set_title("z")
# Histogram
plt.figure()
plt.hist(catalog_in["z"], bins=20)
plt.xlabel("z")
plt.ylabel("count")
# Relation
plt.figure()
plt.scatter(catalog_in["e1"], catalog_in["e2"], s=1.0, alpha=0.2)
plt.xlabel("e1")
plt.ylabel("e2")
make_plots(obs_galaxies)
## 5. Running CLMM on the dataset We use the functions similar to
examples/Paper_v1.0/gt_and_use_case.ipynb.
Make a galaxy cluster object
from clmm import Cosmology
import clmm
cosmo = Cosmology(H0=70.0, Omega_dm0=0.27 - 0.045, Omega_b0=0.045, Omega_k0=0.0)
# We consider RMJ051637.4-543001.6 (ACO S520)
cluster_z = 0.30416065 # Cluster redshift
cluster_ra = 79.155704 # Cluster Ra in deg
cluster_dec = -54.500456 # Cluster Dec in deg
# Select background galaxies
obs_galaxies = obs_galaxies[(obs_galaxies["z"] > (cluster_z + 0.1)) & (obs_galaxies["z"] < 1.5)]
obs_galaxies["id"] = np.arange(len(obs_galaxies))
# Put galaxy values on arrays
gal_ra = obs_galaxies["ra"] # Galaxies Ra in deg
gal_dec = obs_galaxies["dec"] # Galaxies Dec in deg
gal_e1 = obs_galaxies["e1"] # Galaxies elipticipy 1
gal_e2 = obs_galaxies["e2"] # Galaxies elipticipy 2
gal_z = obs_galaxies["z"] # Galaxies observed redshift
gal_id = obs_galaxies["id"] # Galaxies ID
# Create a GCData with the galaxies.
galaxies = clmm.GCData(
[gal_ra, gal_dec, gal_e1, gal_e2, gal_z, gal_id], names=["ra", "dec", "e1", "e2", "z", "id"]
)
# Create a GalaxyCluster.
cluster = clmm.GalaxyCluster("Name of cluster", cluster_ra, cluster_dec, cluster_z, galaxies)
Measure the shear profile
import clmm.dataops as da
# Convert ellipticities into shears for the members.
cluster.compute_tangential_and_cross_components()
print(cluster.galcat.colnames)
# Measure profile and add profile table to the cluster.
cluster.make_radial_profile(
bins=da.make_bins(0.2, 5.0, 7, method="evenlog10width"),
bin_units="Mpc",
cosmo=cosmo,
include_empty_bins=False,
gal_ids_in_bins=True,
)
print(cluster.profile.colnames)
['ra', 'dec', 'e1', 'e2', 'z', 'id', 'theta', 'et', 'ex']
['radius_min', 'radius', 'radius_max', 'gt', 'gt_err', 'gx', 'gx_err', 'z', 'z_err', 'n_src', 'W_l', 'gal_id']
fig, ax = plt.subplots(figsize=(7, 5), ncols=1, nrows=1)
errorbar_kwargs = dict(linestyle="", marker="o", markersize=1, elinewidth=0.5, capthick=0.5)
ax.errorbar(
cluster.profile["radius"],
cluster.profile["gt"],
cluster.profile["gt_err"],
c="k",
**errorbar_kwargs
)
ax.set_xlabel("R [Mpc]", fontsize=10)
ax.set_ylabel(r"$g_t$", fontsize=10)
ax.set_xscale("log")
ax.grid(lw=0.3)
ax.minorticks_on()
ax.grid(which="minor", lw=0.1)
plt.show()
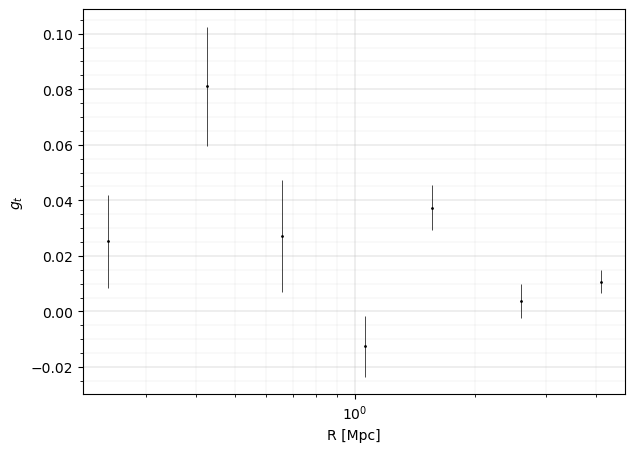
Theoretical predictions
from clmm.utils import convert_units
# Model relying on the overall redshift distribution of the sources (WtG III Applegate et al. 2014).
z_inf = 1000
concentration = 4.0
bs_mean = np.mean(clmm.utils.compute_beta_s(cluster.galcat["z"], cluster_z, z_inf, cosmo))
bs2_mean = np.mean(clmm.utils.compute_beta_s(cluster.galcat["z"], cluster_z, z_inf, cosmo) ** 2)
def predict_reduced_tangential_shear_redshift_distribution(profile, logm):
gt = clmm.compute_reduced_tangential_shear(
r_proj=profile["radius"], # Radial component of the profile
mdelta=10**logm, # Mass of the cluster [M_sun]
cdelta=concentration, # Concentration of the cluster
z_cluster=cluster_z, # Redshift of the cluster
z_src=(bs_mean, bs2_mean), # tuple of (bs_mean, bs2_mean)
z_src_info="beta",
approx="order1",
cosmo=cosmo,
delta_mdef=200,
massdef="critical",
halo_profile_model="nfw",
)
return gt
# Model using individual redshift and radial information, to compute the averaged shear in each radial bin, based on the galaxies actually present in that bin.
cluster.galcat["theta_mpc"] = convert_units(
cluster.galcat["theta"], "radians", "mpc", cluster.z, cosmo
)
def predict_reduced_tangential_shear_individual_redshift(profile, logm):
return np.array(
[
np.mean(
clmm.compute_reduced_tangential_shear(
# Radial component of each source galaxy inside the radial bin
r_proj=cluster.galcat[radial_bin["gal_id"]]["theta_mpc"],
mdelta=10**logm, # Mass of the cluster [M_sun]
cdelta=concentration, # Concentration of the cluster
z_cluster=cluster_z, # Redshift of the cluster
# Redshift value of each source galaxy inside the radial bin
z_src=cluster.galcat[radial_bin["gal_id"]]["z"],
cosmo=cosmo,
delta_mdef=200,
massdef="critical",
halo_profile_model="nfw",
)
)
for radial_bin in profile
]
)
Mass fitting
mask_for_fit = cluster.profile["n_src"] > 2
data_for_fit = cluster.profile[mask_for_fit]
from clmm.support.sampler import fitters
def fit_mass(predict_function):
popt, pcov = fitters["curve_fit"](
predict_function,
data_for_fit,
data_for_fit["gt"],
data_for_fit["gt_err"],
bounds=[10.0, 17.0],
)
logm, logm_err = popt[0], np.sqrt(pcov[0][0])
return {
"logm": logm,
"logm_err": logm_err,
"m": 10**logm,
"m_err": (10**logm) * logm_err * np.log(10),
}
%%time
fit_redshift_distribution = fit_mass(predict_reduced_tangential_shear_redshift_distribution)
fit_individual_redshift = fit_mass(predict_reduced_tangential_shear_individual_redshift)
CPU times: user 258 ms, sys: 1.15 ms, total: 259 ms
Wall time: 259 ms
print(
"Best fit mass for N(z) model ="
f' {fit_redshift_distribution["m"]:.3e} +/- {fit_redshift_distribution["m_err"]:.3e} Msun'
)
print(
"Best fit mass for individual redshift and radius ="
f' {fit_individual_redshift["m"]:.3e} +/- {fit_individual_redshift["m_err"]:.3e} Msun'
)
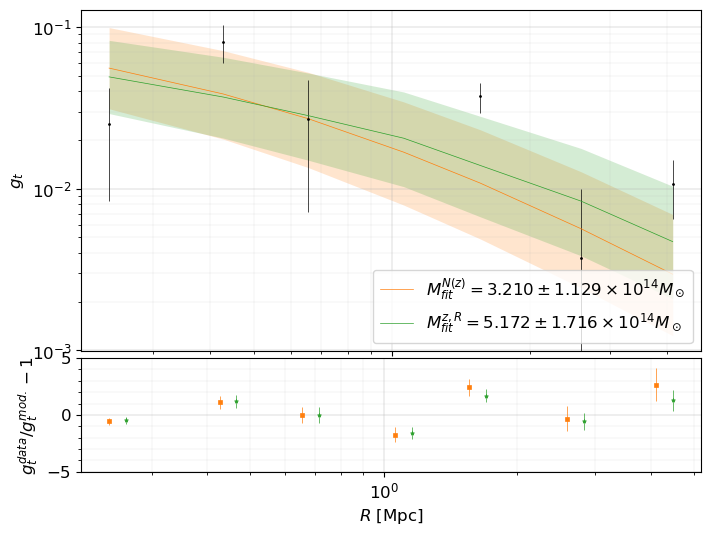
Best fit mass for N(z) model = 3.210e+14 +/- 1.129e+14 Msun
Best fit mass for individual redshift and radius = 5.172e+14 +/- 1.716e+14 Msun
Visualization of the results
def get_predicted_shear(predict_function, fit_values):
gt_est = predict_function(data_for_fit, fit_values["logm"])
gt_est_err = [
predict_function(data_for_fit, fit_values["logm"] + i * fit_values["logm_err"])
for i in (-3, 3)
]
return gt_est, gt_est_err
gt_redshift_distribution, gt_err_redshift_distribution = get_predicted_shear(
predict_reduced_tangential_shear_redshift_distribution, fit_redshift_distribution
)
gt_individual_redshift, gt_err_individual_redshift = get_predicted_shear(
predict_reduced_tangential_shear_individual_redshift, fit_individual_redshift
)
chi2_redshift_distribution_dof = np.sum(
(gt_redshift_distribution - data_for_fit["gt"]) ** 2 / (data_for_fit["gt_err"]) ** 2
) / (len(data_for_fit) - 1)
chi2_individual_redshift_dof = np.sum(
(gt_individual_redshift - data_for_fit["gt"]) ** 2 / (data_for_fit["gt_err"]) ** 2
) / (len(data_for_fit) - 1)
print(f"Reduced chi2 (N(z) model) = {chi2_redshift_distribution_dof}")
print(f"Reduced chi2 (individual (R,z) model) = {chi2_individual_redshift_dof}")
Reduced chi2 (N(z) model) = 4.780165089363783
Reduced chi2 (individual (R,z) model) = 4.411023500984584
fig, axes = plt.subplots(
nrows=2,
ncols=1,
figsize=(8, 6),
gridspec_kw={"height_ratios": [3, 1], "wspace": 0.4, "hspace": 0.03},
)
axes[0].errorbar(
data_for_fit["radius"], data_for_fit["gt"], data_for_fit["gt_err"], c="k", **errorbar_kwargs
)
# Points in grey have not been used for the fit.
axes[0].errorbar(
cluster.profile["radius"][~mask_for_fit],
cluster.profile["gt"][~mask_for_fit],
cluster.profile["gt_err"][~mask_for_fit],
c="grey",
**errorbar_kwargs,
)
pow10 = 14
mlabel = lambda name, fits: (
rf"$M_{{fit}}^{{{name}}} = "
rf'{fits["m"]/10**pow10:.3f}\pm'
rf'{fits["m_err"]/10**pow10:.3f}'
rf"\times 10^{{{pow10}}} M_\odot$"
)
# The model for the 1st method.
axes[0].loglog(
data_for_fit["radius"],
gt_redshift_distribution,
"-C1",
label=mlabel("N(z)", fit_redshift_distribution),
lw=0.5,
)
axes[0].fill_between(
data_for_fit["radius"], *gt_err_redshift_distribution, lw=0, color="C1", alpha=0.2
)
# The model for the 2nd method.
axes[0].loglog(
data_for_fit["radius"],
gt_individual_redshift,
"-C2",
label=mlabel("z,R", fit_individual_redshift),
lw=0.5,
)
axes[0].fill_between(
data_for_fit["radius"], *gt_err_individual_redshift, lw=0, color="C2", alpha=0.2
)
axes[0].set_ylabel(r"$g_t$", fontsize=12)
axes[0].legend(fontsize=12, loc=4)
axes[0].set_xticklabels([])
axes[0].tick_params("x", labelsize=12)
axes[0].tick_params("y", labelsize=12)
axes[1].set_ylim(1.0e-3, 0.5)
errorbar_kwargs2 = {k: v for k, v in errorbar_kwargs.items() if "marker" not in k}
errorbar_kwargs2["markersize"] = 3
errorbar_kwargs2["markeredgewidth"] = 0.5
delta = (cluster.profile["radius"][1] / cluster.profile["radius"][0]) ** 0.15
axes[1].errorbar(
data_for_fit["radius"],
data_for_fit["gt"] / gt_redshift_distribution - 1,
yerr=data_for_fit["gt_err"] / gt_redshift_distribution,
marker="s",
c="C1",
**errorbar_kwargs2,
)
errorbar_kwargs2["markersize"] = 3
errorbar_kwargs2["markeredgewidth"] = 0.5
axes[1].errorbar(
data_for_fit["radius"] * delta,
data_for_fit["gt"] / gt_individual_redshift - 1,
yerr=data_for_fit["gt_err"] / gt_individual_redshift,
marker="*",
c="C2",
**errorbar_kwargs2,
)
axes[1].set_xlabel(r"$R$ [Mpc]", fontsize=12)
axes[1].set_ylabel(r"$g_t^{data}/g_t^{mod.}-1$", fontsize=12)
axes[1].set_xscale("log")
axes[1].set_ylim(-5, 5)
axes[1].tick_params("x", labelsize=12)
axes[1].tick_params("y", labelsize=12)
for ax in axes:
ax.grid(lw=0.3)
ax.minorticks_on()
ax.grid(which="minor", lw=0.1)
plt.show()
# Note since we made cuts on the catalog, the redshift distribution of the remaining sources might not be representative.
References
Zuntz J., Sheldon E., Samuroff S., Troxel M. A., Jarvis M., MacCrann N., Gruen D., et al., 2018, MNRAS, 481, 1149. doi:10.1093/mnras/sty2219
Hoyle B., Gruen D., Bernstein G. M., Rau M. M., De Vicente J., Hartley W. G., Gaztanaga E., et al., 2018, MNRAS, 478, 592. doi:10.1093/mnras/sty957
McClintock T., Varga T. N., Gruen D., Rozo E., Rykoff E. S., Shin T., Melchior P., et al., 2019, MNRAS, 482, 1352. doi:10.1093/mnras/sty2711