Generate mock data for a cluster
In this example we generate mock data with a variety of systematic
effects including photometric redshifts, source galaxy distributions,
and shape noise. We then populate a galaxy cluster object. This
notebooks is organised as follows: - Imports and configuration setup -
Generate mock data with different source galaxy options - Generate mock
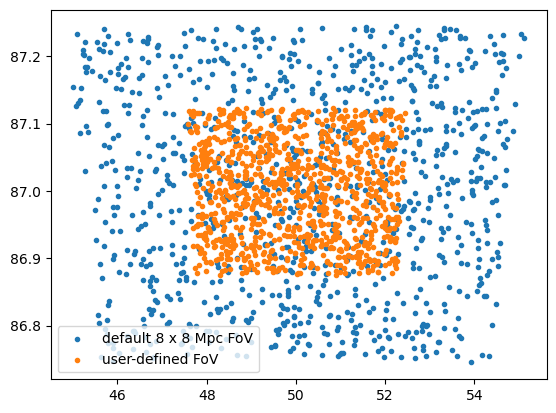
data with different field-of-view options - Generate mock data with
different galaxy cluster options (only available with the Numcosmo
and/or CCL backends). Use the os.environ['CLMM_MODELING_BACKEND']
line below to select your backend.
import os
## Uncomment the following line if you want to use a specific modeling backend among 'ct' (cluster-toolkit), 'ccl' (CCL) or 'nc' (Numcosmo). Default is 'ccl'
# os.environ['CLMM_MODELING_BACKEND'] = 'nc'
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import clmm
Make sure we know which version we’re using
clmm.__version__
'1.12.0'
Import mock data module and setup the configuration
from clmm.support import mock_data as mock
from clmm import Cosmology
Mock data generation requires a defined cosmology
mock_cosmo = Cosmology(H0=70.0, Omega_dm0=0.27 - 0.045, Omega_b0=0.045, Omega_k0=0.0)
Mock data generation requires some cluster information. The default is to work with the NFW profile, using the “200,mean” mass definition. The Numcosmo and CCL backends allow for more flexibility (see last section of this notebook)
cosmo = mock_cosmo
cluster_id = "Awesome_cluster"
cluster_m = 1.0e15 # M200,m
cluster_z = 0.3
src_z = 0.8
concentration = 4
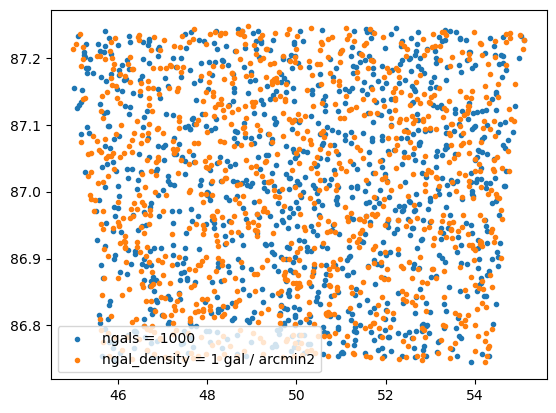
ngals = 1000 # number of source galaxies
# Cluster centre coordinates
cluster_ra = 50.0
cluster_dec = 87.0
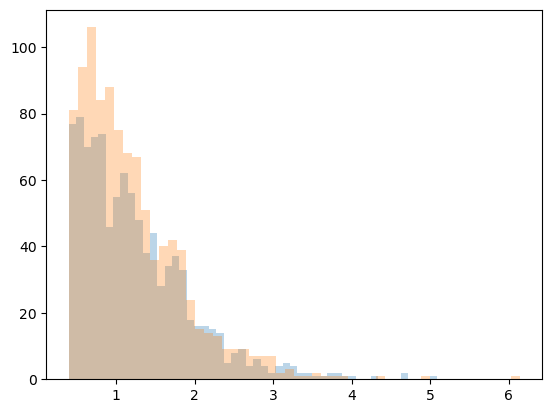
Generate the mock catalog with different source galaxy options
Clean data: no noise, all galaxies at the same redshift
zsrc_min = cluster_z + 0.1
ideal_data = mock.generate_galaxy_catalog(
cluster_m,
cluster_z,
concentration,
cosmo,
src_z,
ngals=ngals,
cluster_ra=cluster_ra,
cluster_dec=cluster_dec,
)
# let's put all these quantities in a single dictionary to facilitate clarity
cluster_kwargs = {
"cluster_m": cluster_m,
"cluster_z": cluster_z,
"cluster_ra": cluster_ra,
"cluster_dec": cluster_dec,
"cluster_c": concentration,
"cosmo": cosmo,
}
ideal_data = mock.generate_galaxy_catalog(**cluster_kwargs, zsrc=src_z, ngals=ngals)
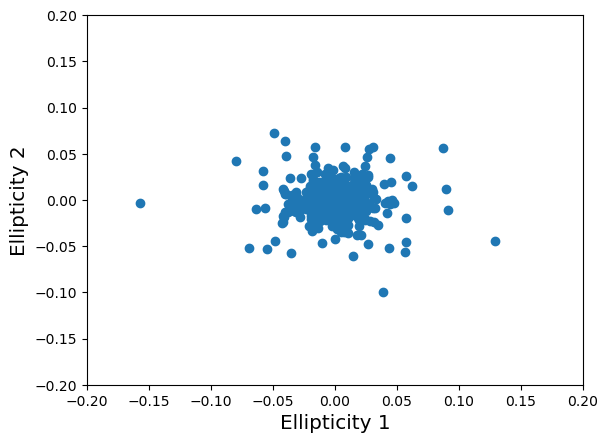
Noisy data: shape noise, all galaxies at the same redshift
noisy_data_src_z = mock.generate_galaxy_catalog(
**cluster_kwargs, zsrc=src_z, shapenoise=0.05, ngals=ngals
)
Noisy data: shape noise plus measurement error, all galaxies at the same redshift
noisy_data_src_z_e_err = mock.generate_galaxy_catalog(
**cluster_kwargs, zsrc=src_z, shapenoise=0.05, mean_e_err=0.05, ngals=ngals
)
WARNING: Experimental feature. Uncertainties are created by
simply drawing random numbers near the value specified by
mean_e_err. Use at your own risk. This will be improved in future
releases.
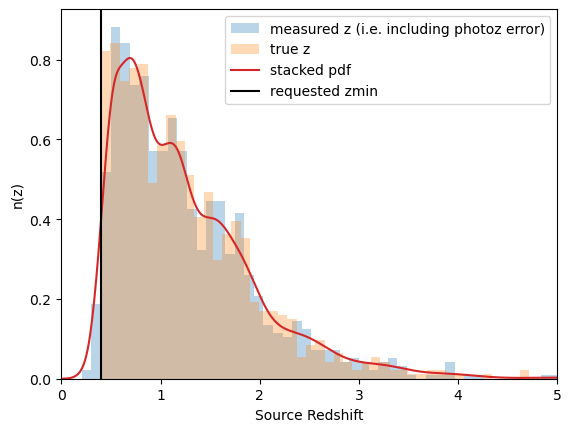
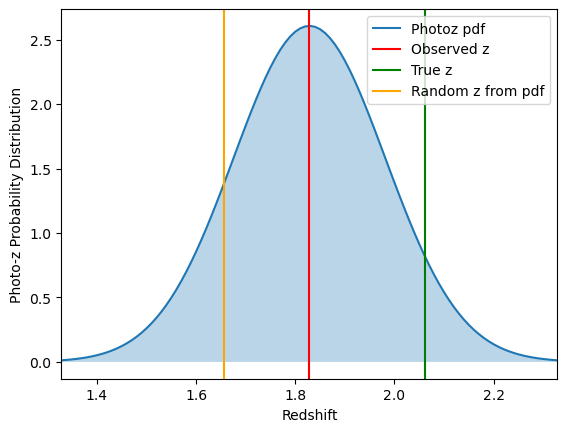
Noisy data: photo-z errors (and pdfs!), all galaxies at the same redshift. At present, the pdfs are generated by drawing a random value from a Normal distribution with mean
ztrueand width given bydz = (1+z)*photoz_sigma_unscaled, and the pdf is this Normal distribution centered aroundzinstead ofztrue.
noisy_data_photoz = mock.generate_galaxy_catalog(
**cluster_kwargs,
zsrc=src_z,
shapenoise=0.05,
photoz_sigma_unscaled=0.05,
pz_bins=101,
ngals=ngals
)
In the default PDF storing (pzpdf_type='shared_bins'), the values of
the PDF are added to the pzpdf column and the binning scheme is
stored in the pzpdf_info attribute:
noisy_data_photoz.pzpdf_info
{'type': 'shared_bins',
'zbins': array([0. , 0.01959865, 0.0391973 , 0.05879595, 0.0783946 ,
0.09799325, 0.1175919 , 0.13719055, 0.15678921, 0.17638786,
0.19598651, 0.21558516, 0.23518381, 0.25478246, 0.27438111,
0.29397976, 0.31357841, 0.33317706, 0.35277571, 0.37237436,
0.39197301, 0.41157166, 0.43117031, 0.45076896, 0.47036762,
0.48996627, 0.50956492, 0.52916357, 0.54876222, 0.56836087,
0.58795952, 0.60755817, 0.62715682, 0.64675547, 0.66635412,
0.68595277, 0.70555142, 0.72515007, 0.74474872, 0.76434737,
0.78394603, 0.80354468, 0.82314333, 0.84274198, 0.86234063,
0.88193928, 0.90153793, 0.92113658, 0.94073523, 0.96033388,
0.97993253, 0.99953118, 1.01912983, 1.03872848, 1.05832713,
1.07792578, 1.09752444, 1.11712309, 1.13672174, 1.15632039,
1.17591904, 1.19551769, 1.21511634, 1.23471499, 1.25431364,
1.27391229, 1.29351094, 1.31310959, 1.33270824, 1.35230689,
1.37190554, 1.3915042 , 1.41110285, 1.4307015 , 1.45030015,
1.4698988 , 1.48949745, 1.5090961 , 1.52869475, 1.5482934 ,
1.56789205, 1.5874907 , 1.60708935, 1.626688 , 1.64628665,
1.6658853 , 1.68548395, 1.70508261, 1.72468126, 1.74427991,
1.76387856, 1.78347721, 1.80307586, 1.82267451, 1.84227316,
1.86187181, 1.88147046, 1.90106911, 1.92066776, 1.94026641,
1.95986506])}
noisy_data_photoz[:5]
defined by: cosmo=None
with columns: ra, dec, e1, e2, z, ztrue, pzpdf, id
and pzpdf: shared_bins [0. 0.02 0.04 0.06 0.08 ... 1.88 1.9 1.92 1.94 1.96]
5 objects
| ra | dec | e1 | e2 | z | ztrue | pzpdf | id |
|---|---|---|---|---|---|---|---|
| float64 | float64 | float64 | float64 | float64 | float64 | float64[101] | int64 |
| 46.25728808233552 | 86.8922305424305 | 0.08236207803522719 | -0.05411854076714888 | 0.6987103181997064 | 0.8 | 3.622089304592751e-13 .. 1.0180322952717169e-42 | 0 |
| 50.07778715623189 | 87.10740150490655 | -0.01475225297133639 | 0.03882800844119587 | 0.7392962132175859 | 0.8 | 9.870794333079776e-15 .. 5.1047370592912794e-40 | 1 |
| 48.40809879885119 | 86.87728907171557 | -1.2575884722892804e-05 | 0.02096267636736204 | 0.9009303274293756 | 0.8 | 7.709520397996465e-22 .. 3.849218150728941e-30 | 2 |
| 53.014368911831866 | 86.9440239760556 | -0.011911350886457556 | -0.05893011144277411 | 0.8447969829350334 | 0.8 | 3.2663820149084725e-19 .. 2.0599039175494822e-33 | 3 |
| 53.435230187909205 | 87.09017457767816 | -0.01999543117339179 | 0.02354367773663435 | 0.6191383565693703 | 0.8 | 2.345198551898936e-10 .. 2.86718907682299e-48 | 4 |