Generate mock data for a cluster ensemble
Generate cluster ensemble with random masses and redshifts
%load_ext autoreload
%autoreload 2
import sys
import os
import numpy as np
from astropy.table import Table
from numpy import random
import scipy
import matplotlib.pyplot as plt
import clmm
from clmm import GalaxyCluster, ClusterEnsemble, GCData
from clmm import Cosmology
from clmm.support import mock_data as mock
clmm.__version__
'1.12.0'
np.random.seed(11)
cosmo = Cosmology(H0=71.0, Omega_dm0=0.265 - 0.0448, Omega_b0=0.0448, Omega_k0=0.0)
Generating a cluster catalog and associated source catalogs
Below, we randomly generate the masses, redshifts, concentrations and
coordinates of an ensemble of n_clusters clusters. For simplicity,
the drawing is performed uniformly in logm and redshift (instead of
following a halo mass function).
# redshift and mass range of the galaxy clusters
z_bin = [0.2, 0.25]
logm_bin = np.array([14, 14.1]) # Solar Mass
# number of clusters in the ensemble
n_clusters = 30
# random draw in the mass and redshift range (for simplicity, uniform instead of following an actual mass function)
cluster_m = 10 ** (
(logm_bin[1] - logm_bin[0]) * np.random.random(n_clusters) + logm_bin[0]
) # in M_sun
cluster_z = (z_bin[1] - z_bin[0]) * np.random.random(n_clusters) + z_bin[0]
# random normal draw of cluster concentration, around c_mean
c_mean = 4.0
lnc = abs(np.log(c_mean) + 0.01 * np.random.randn(n_clusters))
concentration = np.exp(lnc)
# randomly draw cluster positions on the sky
ra = np.random.random(n_clusters) * 360 # from 0 to 360 deg
sindec = np.random.random(n_clusters) * 2 - 1
dec = np.arcsin(sindec) * 180 / np.pi # from -90 to 90 deg
Background galaxy catalog generation
For each cluster of the ensemble, we use mock_data to generate a
background galaxy catalog and store the results in a GalaxyCluster
object. Note that: - The cluster density profiles follow the NFW
parametrisation; - The source redshifts follow the Chang et
al. distribution and have associated pdfs; - The shapes include shape
noise and shape measurement errors; - Background galaxy catalogs are
independent, even if the clusters are close (i.e., no common galaxy
between two catalogs). - For each cluster we then compute - the
tangential and cross \(\Delta\Sigma\) for each background galaxy -
the weights w_ls to be used to compute the corresponding radial
profiles (see demo_compute_deltasigma_weights.ipynb notebook for
details)
The cluster objects are then stored in gclist.
import warnings
warnings.simplefilter(
"ignore"
) # just to prevent warning print out when looping over the cluster ensemble below
gclist = []
# number of galaxies in each cluster field (alternatively, can use the galaxy density instead)
n_gals = 10000
# ngal_density = 10
for i in range(n_clusters):
# generate background galaxy catalog for cluster i
noisy_data_z = mock.generate_galaxy_catalog(
cluster_m[i],
cluster_z[i],
concentration[i],
cosmo,
cluster_ra=ra[i],
cluster_dec=dec[i],
zsrc="chang13",
delta_so=200,
massdef="critical",
halo_profile_model="nfw",
zsrc_min=cluster_z[i] + 0.1,
zsrc_max=3.0,
field_size=10.0,
shapenoise=0.04,
photoz_sigma_unscaled=0.02,
ngals=n_gals,
mean_e_err=0.1,
)
cl = clmm.GalaxyCluster("mock_cluster", ra[i], dec[i], cluster_z[i], noisy_data_z)
# compute DeltaSigma for each background galaxy
cl.compute_tangential_and_cross_components(
shape_component1="e1",
shape_component2="e2",
tan_component="DS_t",
cross_component="DS_x",
cosmo=cosmo,
is_deltasigma=True,
use_pdz=True,
)
# compute the weights to be used to bluid the DeltaSigma radial profiles
cl.compute_galaxy_weights(
use_pdz=True,
use_shape_noise=True,
shape_component1="e1",
shape_component2="e2",
use_shape_error=True,
shape_component1_err="e_err",
shape_component2_err="e_err",
weight_name="w_ls",
cosmo=cosmo,
is_deltasigma=True,
add=True,
)
# append the cluster in the list
gclist.append(cl)
cl.galcat.columns
<TableColumns names=('ra','dec','e1','e2','e_err','z','ztrue','pzpdf','id','sigma_c_eff','theta','DS_t','DS_x','w_ls')>
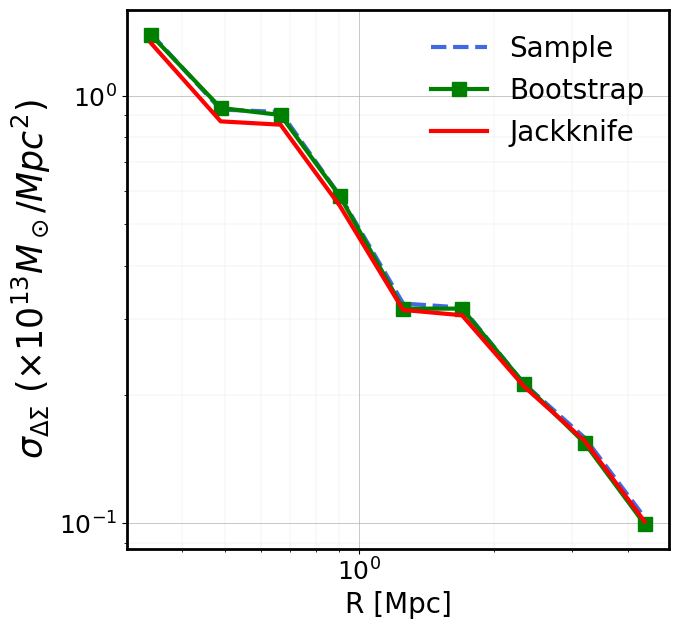
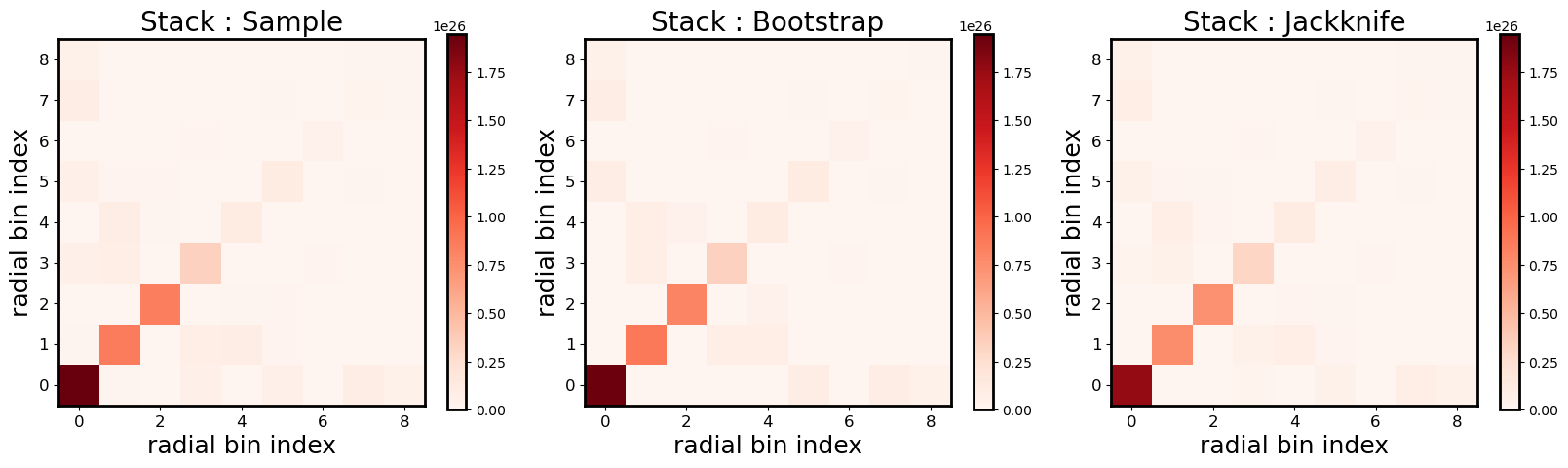
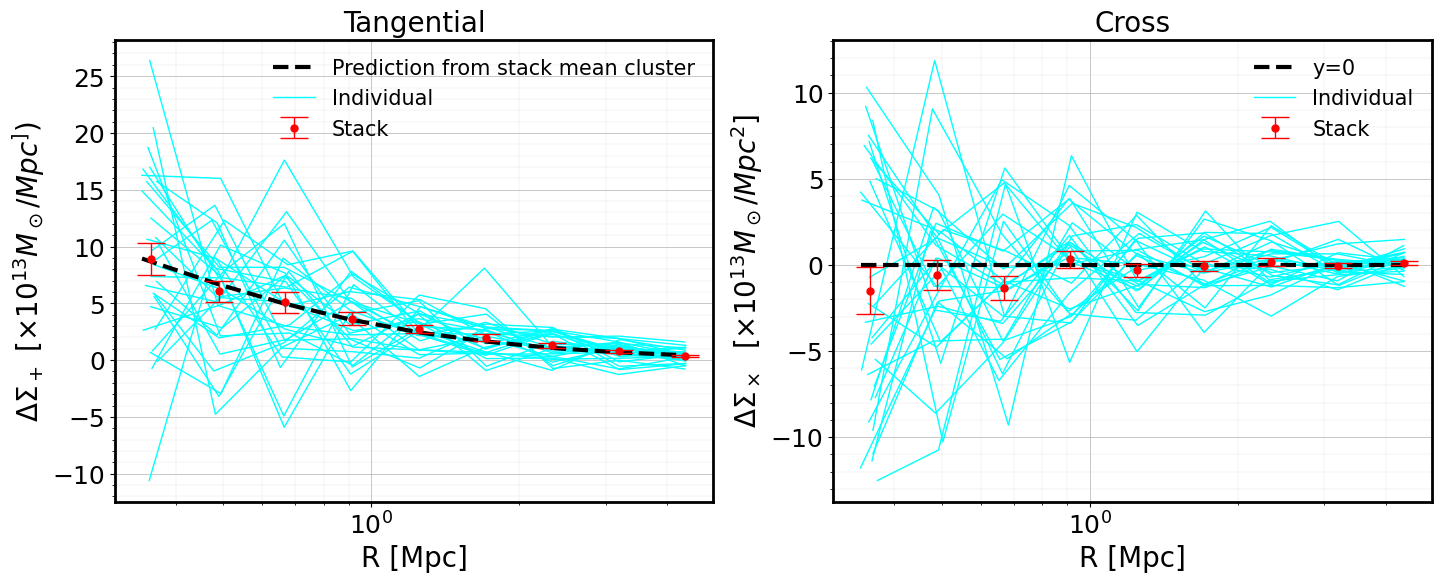
Creating ClusterEnsemble object and estimation of individual excess surface density profiles
From the galaxy cluster object list gclist, we instantiate a cluster
ensemble object clusterensemble. This instantiation step uses - the
individual galaxy input \(\Delta\Sigma_+\) and
\(\Delta\Sigma_{\times}\) values (computed in the previous step,
DS_{t,x}) - the corresponding individual input weights w_ls
(computed in the previous step)
to compute
the output tangential
DS_tand cross signalDS_xbinned profiles (where the binning is controlled bybins)the associated profile weights
W_l(that will be used to compute the stacked profile)
ensemble_id = 1
names = ["id", "ra", "dec", "z", "radius", "gt", "gx", "W_l"]
bins = np.logspace(np.log10(0.3), np.log10(5), 10)
clusterensemble = ClusterEnsemble(
ensemble_id,
gclist,
tan_component_in="DS_t",
cross_component_in="DS_x",
tan_component_out="DS_t",
cross_component_out="DS_x",
weights_in="w_ls",
weights_out="W_l",
bins=bins,
bin_units="Mpc",
cosmo=cosmo,
)
There is also the option to create an ClusterEnsemble object without
the clusters list. Then, the user may compute the individual profile for
each wanted cluster and compute the radial profile once all the
indvidual profiles have been computed. This method may be reccomended if
there a large number of clusters to avoid excess of memory allocation,
since the user can generate each cluster separately, compute its
individual profile and then delete the cluster object.
ensemble_id2 = 2
empty_cluster_ensemble = ClusterEnsemble(ensemble_id2)
for cluster in gclist:
empty_cluster_ensemble.make_individual_radial_profile(
galaxycluster=cluster,
tan_component_in="DS_t",
cross_component_in="DS_x",
tan_component_out="DS_t",
cross_component_out="DS_x",
weights_in="w_ls",
weights_out="W_l",
bins=bins,
bin_units="Mpc",
cosmo=cosmo,
)
A third option is where all clusters already have the profile computed,
and we just add those to the ClusterEnsemble object:
# add profiles to gclist
for cluster in gclist:
cluster.make_radial_profile(
tan_component_in="DS_t",
cross_component_in="DS_x",
tan_component_out="DS_t",
cross_component_out="DS_x",
weights_in="w_ls",
weights_out="W_l",
bins=bins,
bin_units="Mpc",
cosmo=cosmo,
use_weights=True,
)
cluster.profile
defined by: cosmo='CCLCosmology(H0=71.0, Omega_dm0=0.2202, Omega_b0=0.0448, Omega_k0=0.0)', bin_units='Mpc'
with columns: radius_min, radius, radius_max, DS_t, DS_t_err, DS_x, DS_x_err, z, z_err, n_src, W_l
9 objects
| radius_min | radius | radius_max | DS_t | DS_t_err | DS_x | DS_x_err | z | z_err | n_src | W_l |
|---|---|---|---|---|---|---|---|---|---|---|
| float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | int64 | float64 |
| 0.29999999999999993 | 0.3553888808485904 | 0.4100928778589602 | 47114109394812.89 | 74981653954774.03 | 71725831060887.97 | 70821817988064.0 | 1.2294188552708238 | 0.1254222511654474 | 29 | 6.36216161623601e-29 |
| 0.4100928778589602 | 0.4971275421239857 | 0.5605872282354804 | 28407616591812.977 | 42764109085226.86 | -57071960817478.72 | 46736076941033.97 | 1.377144531885036 | 0.09702952618749792 | 49 | 1.15161521880834e-28 |
| 0.5605872282354804 | 0.6712361227940055 | 0.7663094323935532 | 74964576073867.19 | 38525941380754.64 | 56308907913134.02 | 42228781968987.93 | 1.4005388323178487 | 0.06709616391264094 | 86 | 2.113338478541802e-28 |
| 0.7663094323935532 | 0.9063818034287909 | 1.0475268015357952 | 31222548973785.86 | 28953248329270.203 | 4403827566750.543 | 30962934549207.242 | 1.357185274715219 | 0.054794861435771995 | 158 | 3.639913884168516e-28 |
| 1.0475268015357952 | 1.2573762610803276 | 1.4319442689206874 | 43299578835108.54 | 21990385325328.957 | 150722770699.83386 | 20759010434230.527 | 1.3451641041042115 | 0.03497735694036869 | 296 | 7.038287482539166e-28 |
| 1.4319442689206874 | 1.7043857764046686 | 1.9574338205844326 | 9749022992700.264 | 15940514809587.195 | -8821468381838.701 | 15585239988553.379 | 1.3306378739615503 | 0.02681950361641807 | 535 | 1.270287908236562e-27 |
| 1.9574338205844326 | 2.339049853316992 | 2.6757655623397656 | 26294486206933.47 | 10815129177769.555 | -29746134079554.895 | 10982086708852.97 | 1.3595192146604755 | 0.01957031696068926 | 1051 | 2.521188293741188e-27 |
| 2.6757655623397656 | 3.194399428042595 | 3.6577079997860453 | 4645975281894.578 | 7782221363303.094 | -6755554338437.702 | 7744746597065.146 | 1.3679035365281018 | 0.014534053109116193 | 1951 | 4.719527212638533e-27 |
| 3.6577079997860453 | 4.368455348815572 | 5.000000000000001 | 3195312396468.3667 | 5740366240659.977 | -1692230857239.375 | 5890154465299.272 | 1.3496089726931957 | 0.010516912111932255 | 3644 | 8.730103338179242e-27 |