Generate “realistic” mock data for a cluster ensemble
Generate cluster ensemble with “realistic” mass and redshift distribution
In this notebook, we generate a ‘realistic’ mock cluster ensemble, i.e. given a halo mass function, and mass-concentration relation (more complexity, such as miscentering, could also be added but it’s not done here). The notebook is structured as follows: - Implementation of the CCL and NumCosmo mass functions. - Some comparisons between the different backends. - Implementation of a accept-reject method to generate cluster samples from the mass function. - Generation of a cluster catalog for mass and redshift bins followed by a comparison with the theoretical prediction
Then, the remaining of the notebook repeats what was done in the
Example_cluster_ensemble.ipynb. - Geration of mock galaxy data for
each cluster - Creating ClusterEnsemble object and estimation of
individual excess surface density profiles - An analysis of the
covariance of the stack between radial bins
Draw pairs of mass and redshift according to a mass function (computed with either CCL or NumCosmo)
In this notebook we use the Tinker halo mass function, and demonstrate the implementation via CCL or NumCosmo.
import sys
import numpy as np
import pyccl as ccl
import matplotlib.pyplot as plt
import scipy
import clmm
from clmm import GalaxyCluster, ClusterEnsemble, GCData
from clmm import Cosmology
from clmm.support import mock_data as mock
%matplotlib inline
# NumCosmo imports
try:
import gi
gi.require_version("NumCosmo", "1.0")
gi.require_version("NumCosmoMath", "1.0")
except:
pass
from gi.repository import GObject
from gi.repository import NumCosmo as Nc
from gi.repository import NumCosmoMath as Ncm
Ncm.cfg_init()
Ncm.cfg_set_log_handler(lambda msg: sys.stdout.write(msg) and sys.stdout.flush())
For reproducibility:
np.random.seed(11)
Mass function - CCL implementation
cosmo = ccl.Cosmology(
Omega_c=0.26,
Omega_b=0.04,
h=0.7,
sigma8=0.8,
n_s=0.96,
Neff=3.04,
m_nu=1.0e-05,
mass_split="single",
)
hmd_200c = ccl.halos.MassDef(200, "critical")
# For a different multiplicty function, the user must change this function below
def tinker08_ccl(logm, z):
mass = 10 ** (logm)
hmf_200c = ccl.halos.MassFuncTinker08(mass_def=hmd_200c)
nm = hmf_200c(cosmo, mass, 1.0 / (1 + z))
return nm # dn/dlog10M
# Computing the volume element
def dV_over_dOmega_dz(z):
a = 1.0 / (1.0 + z)
da = ccl.background.angular_diameter_distance(cosmo, a)
E = ccl.background.h_over_h0(cosmo, a)
return ((1.0 + z) ** 2) * (da**2) * ccl.physical_constants.CLIGHT_HMPC / cosmo["h"] / E
def pdf_tinker08_ccl(logm, z):
return tinker08_ccl(logm, z) * dV_over_dOmega_dz(z)
Mass function - Numcosmo implementation
Ncm.cfg_init()
# cosmo_nc = Nc.HICosmoDEXcdm()
cosmo_nc = Nc.HICosmo.new_from_name(Nc.HICosmo, "NcHICosmoDECpl{'massnu-length':<1>}")
cosmo_nc.omega_x2omega_k()
cosmo_nc.param_set_by_name("w0", -1.0)
cosmo_nc.param_set_by_name("w1", 0.0)
cosmo_nc.param_set_by_name("Tgamma0", 2.725)
cosmo_nc.param_set_by_name("massnu_0", 0.0)
cosmo_nc.param_set_by_name("H0", 70.0)
cosmo_nc.param_set_by_name("Omegab", 0.04)
cosmo_nc.param_set_by_name("Omegac", 0.26)
cosmo_nc.param_set_by_name("Omegak", 0.0)
# ENnu = 3.046 - 3.0 * \
# cosmo_nc.E2Press_mnu(1.0e10) / (cosmo_nc.E2Omega_g(1.0e10)
# * (7.0/8.0*(4.0/11.0)**(4.0/3.0)))
ENnu = 3.046
cosmo_nc.param_set_by_name("ENnu", ENnu)
reion = Nc.HIReionCamb.new()
prim = Nc.HIPrimPowerLaw.new()
cosmo_nc.add_submodel(reion)
cosmo_nc.add_submodel(prim)
dist = Nc.Distance.new(2.0)
dist.prepare_if_needed(cosmo_nc)
tf = Nc.TransferFunc.new_from_name("NcTransferFuncEH")
psml = Nc.PowspecMLTransfer.new(tf)
psml.require_kmin(1.0e-6)
psml.require_kmax(1.0e3)
psf = Ncm.PowspecFilter.new(psml, Ncm.PowspecFilterType.TOPHAT)
psf.set_best_lnr0()
prim.props.n_SA = 0.96
old_amplitude = np.exp(prim.props.ln10e10ASA)
prim.props.ln10e10ASA = np.log((0.8 / cosmo_nc.sigma8(psf)) ** 2 * old_amplitude)
print(0.8, cosmo_nc.sigma8(psf))
mulf = Nc.MultiplicityFuncTinker.new()
mulf.set_mdef(Nc.MultiplicityFuncMassDef.CRITICAL)
mulf.set_Delta(200.0)
mf = Nc.HaloMassFunction.new(dist, psf, mulf)
#
# New mass function object using the objects defined above.
#
def tinker08_nc(logm, z):
lnm = logm * np.log(10.0) # convert log10(M) to ln(M)
res = mf.dn_dlnM(cosmo_nc, lnm, z)
return res * np.log(10.0) # convert dn/dlnM to dn/dlog10M
def pdf_tinker08_nc(logm, z):
return tinker08_nc(logm, z) * mf.dv_dzdomega(cosmo_nc, z)
0.8 0.8
Sanity Checks - compare the CCL and NC implementations
Volume element
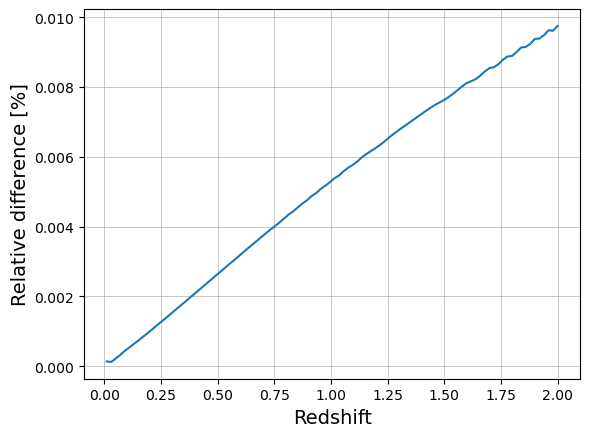
z_arr = np.linspace(0.01, 2, 100)
v_nc = [mf.dv_dzdomega(cosmo_nc, z) for z in z_arr]
v_ccl = dV_over_dOmega_dz(z_arr)
plt.plot(z_arr, np.abs(v_nc / v_ccl - 1) * 100)
plt.xlabel("Redshift", size=14)
plt.ylabel("Relative difference [%]", size=14)
plt.grid(which="major", lw=0.5)
plt.grid(which="minor", lw=0.2)
Mass function
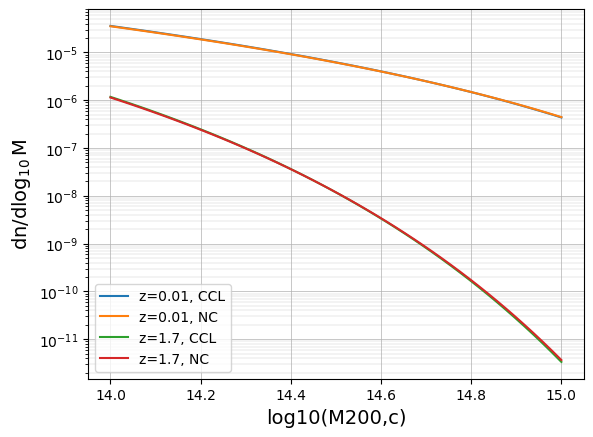
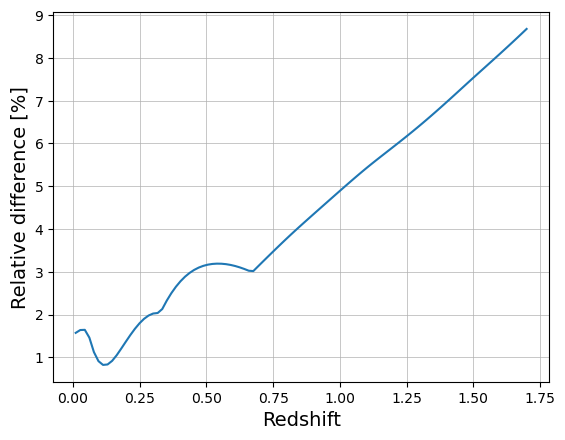
The CCL and Numcosmo implementations give different results for the Tinker08 HMF, especially at high redshift (>1.5). Need to understand why, although it is not a regime we should find ourselves in with cluster studies at optical wavelength. Might be that the CCL and NC cosmologies defined above are not actually the same (neutrinos?).
logm_arr = np.linspace(14.0, 15)
z_arr = np.linspace(0.01, 1.7, 2)
t08_vec_ccl = np.vectorize(tinker08_ccl)
t08_vec_nc = np.vectorize(tinker08_nc)
for z in z_arr:
res_ccl = t08_vec_ccl(logm_arr, z)
res_nc = t08_vec_nc(logm_arr, z)
plt.plot(logm_arr, res_ccl, label=f"z={z}, CCL")
plt.plot(logm_arr, res_nc, label=f"z={z}, NC")
plt.yscale("log")
plt.xlabel("log10(M200,c)", size=14)
plt.ylabel("dn/d$\log_{10}$M", size=14)
plt.legend()
plt.grid(which="major", lw=0.5)
plt.grid(which="minor", lw=0.2)
z_arr = np.linspace(0.01, 1.7, 100)
logm = 15.0
res_ccl = t08_vec_ccl(logm, z_arr)
res_nc = t08_vec_nc(logm, z_arr)
plt.plot(z_arr, np.abs(res_ccl / res_nc - 1) * 100)
plt.xlabel("Redshift", size=14)
plt.ylabel("Relative difference [%]", size=14)
plt.grid(which="major", lw=0.5)
plt.grid(which="minor", lw=0.2)
Acceptance-rejection method to sample (M,z) from the mass function
def bivariate_draw(
pdf, N=1000, logm_min=14.0, logm_max=15.0, zmin=0.01, zmax=1, Ngrid_m=30, Ngrid_z=30
):
"""
Uses the rejection method for generating random numbers derived from an arbitrary
probability distribution.
Parameters
----------
pdf : func
2d distribution function to sample
N : int
number of points to generate
log_min,logm_max : float
log10 mass range
zmin,zmax : float
redshift range
Returns:
--------
ran_logm : list
accepted logm values
ran_z : list
accepted redshift values
acceptance : float
acceptance ratio of the method
"""
# maximum value of the pdf over the mass and redshift space.
# the pdf is not monotonous in mass and redshift, so we need
# to find the maximum numerically. Here we scan the space
# with a regular grid and use the maximum value.
# Accuracy of the results depends on Ngrid_m and Ngrid_z
# This should probably be improved
logM_arr = np.linspace(logm_min, logm_max, Ngrid_m)
z_arr = np.logspace(np.log10(zmin), np.log10(zmax), Ngrid_z)
p = []
for logM in logM_arr:
for z in z_arr:
p.append(pdf(logM, z))
pmax = np.max(p)
pmin = np.min(p)
# Counters
naccept = 0
ntrial = 0
# Keeps drawing until N points are accepted
ran_logm = [] # output list of random numbers
ran_z = [] # output list of random numbers
while naccept < N:
# draw (logm,z) from uniform distribution
# draw p from uniform distribution
logm = np.random.uniform(logm_min, logm_max) # x'
z = np.random.uniform(zmin, zmax) # x'
p = np.random.uniform(0.0, pmax) # y'
if p < pdf(logm, z):
# keep the point
# print(logm, z, p, pdf(logm, z))
ran_logm.append(logm)
ran_z.append(z)
naccept = naccept + 1
ntrial = ntrial + 1
ran_logm = np.asarray(ran_logm)
ran_z = np.asarray(ran_z)
acceptance = float(N / ntrial)
print(f"acceptance = {acceptance}")
return ran_logm, ran_z, acceptance
Draw \(10^4\) (M,z) pairs and check consistency with mass function
# Define the numbers of pairs to draw and the mass and redshift ranges
N = 10000
logm_min = 14.0
logm_max = 14.5
zmin = 0.2
zmax = 1.0
# Choose between the CCL and NC implementation (NC is faster)
# pdf = pdf_tinker08_ccl
pdf = pdf_tinker08_nc
# Normalisation factor for the pdf to be used later
norm = (
1.0
/ scipy.integrate.dblquad(
pdf, zmin, zmax, lambda x: logm_min, lambda x: logm_max, epsrel=1.0e-4
)[0]
)
# Random draw
ran_logm, ran_z, acceptance = bivariate_draw(
pdf, N=N, logm_min=logm_min, logm_max=logm_max, zmin=zmin, zmax=zmax
)
acceptance = 0.32901230506020923
plt.hist2d(ran_logm, ran_z, bins=10)
pass
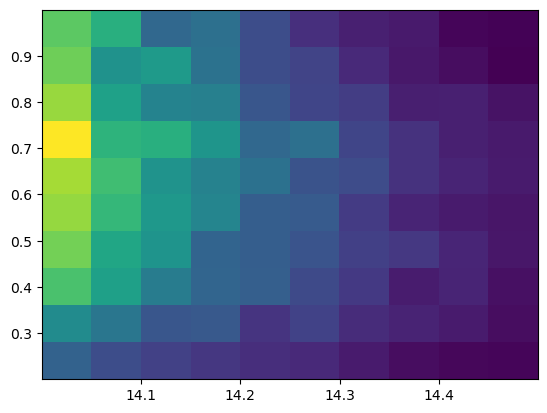
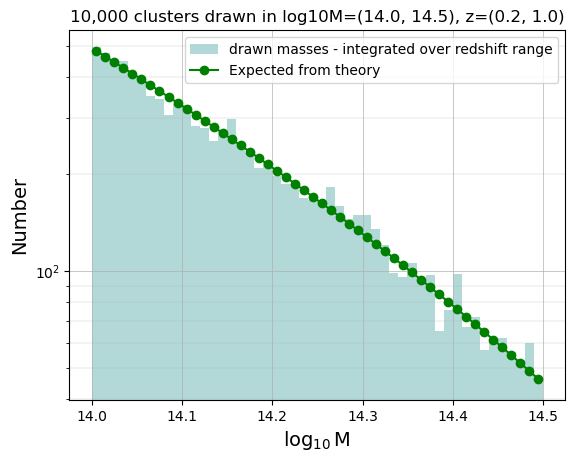
Check distribution in mass bins (integrate over the full redshift range)
hist_m = np.histogram(ran_logm, bins=50)
print(zmin, zmax)
predicted_T08_m_quad = [
norm
* N
* scipy.integrate.dblquad(pdf, zmin, zmax, lambda x: lmmin, lambda x: lmmax, epsrel=1.0e-4)[0]
for lmmin, lmmax in zip(hist_m[1], hist_m[1][1:])
]
0.2 1.0
bin_centers_m = 0.5 * (hist_m[1][:-1] + hist_m[1][1:])
plt.hist(
ran_logm,
bins=hist_m[1],
color="teal",
alpha=0.3,
label="drawn masses - integrated over redshift range",
)
plt.plot(
bin_centers_m, np.array(predicted_T08_m_quad), "-o", color="green", label="Expected from theory"
)
plt.yscale("log")
plt.xlabel("$\log_{10}$M", size=14)
plt.ylabel("Number", size=14)
plt.title(f"{N:,} clusters drawn in log10M={logm_min, logm_max}, z={zmin, zmax}")
plt.legend()
plt.grid(which="major", lw=0.5)
plt.grid(which="minor", lw=0.2)
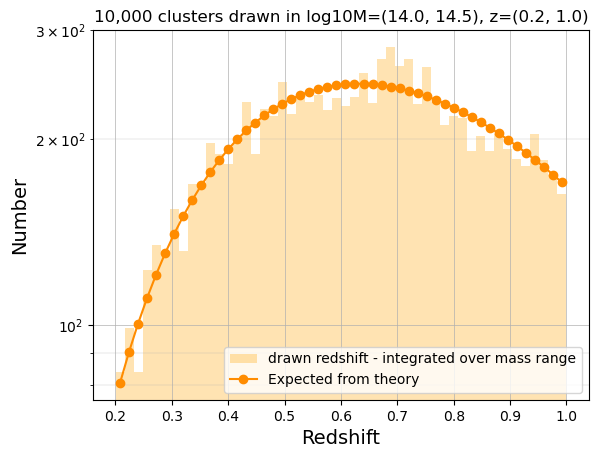
Check distribution in redshift bins (integrate over the full mass range)
hist_z = np.histogram(ran_z, bins=50)
print(logm_min, logm_max)
predicted_T08_z_quad = [
norm
* N
* scipy.integrate.dblquad(
pdf, zzmin, zzmax, lambda x: logm_min, lambda x: logm_max, epsrel=1.0e-4
)[0]
for zzmin, zzmax in zip(hist_z[1], hist_z[1][1:])
]
14.0 14.5
bin_centers_z = 0.5 * (hist_z[1][:-1] + hist_z[1][1:])
plt.hist(
ran_z,
bins=hist_z[1],
color="orange",
alpha=0.3,
label="drawn redshift - integrated over mass range",
)
plt.plot(
bin_centers_z,
np.array(predicted_T08_z_quad),
"-o",
color="darkorange",
label="Expected from theory",
)
plt.yscale("log")
plt.xlabel("Redshift", size=14)
plt.ylabel("Number", size=14)
plt.title(f"{N:,} clusters drawn in log10M={logm_min, logm_max}, z={zmin, zmax}")
plt.legend(loc=4)
plt.grid(which="major", lw=0.5)
plt.grid(which="minor", lw=0.2)
Generating a cluster catalog and associated source catalogs
First, set the cluster masses and redshifts given the generated mock data. Only keep 30 clusters from N generated above to avoid memory issues.
Then, instantiate a CCL concentration object to compute the concentration for each cluster from a mass-concentration realtion (Duffy et al. 2008). The actual concentration for each cluster is drawn from a lognormal distribution around the mean value
Last, we randomly generate the cluster center coordinates over the full sky (from 0 to 360 deg for ra and from -90 to 90 deg to dec)
Cluster ensemble masses, redshifts and positions
# Randomly keep 30 (M,z) pairs from the 1000s generated abve.
Nstack = 30
indices = np.random.choice(len(ran_logm), Nstack)
cluster_m = 10 ** ran_logm[indices]
cluster_z = ran_z[indices]
# Concentration CCL object to compute the theoretical concentration
conc_obj = ccl.halos.ConcentrationDuffy08(mass_def=hmd_200c)
conc_list = []
for number in range(0, len(cluster_m)):
a = 1.0 / (1.0 + (cluster_z[number]))
# mean value of the concentration for that cluster
lnc_mean = np.log(conc_obj(cosmo, M=(cluster_m[number]), a=a))
# random draw of actual concentration from normal distribution around lnc_mean, with a 0.14 scatter
lnc = np.random.normal(lnc_mean, 0.14)
conc_list.append(np.exp(lnc))
conc_list = np.array(conc_list)
# randomly draw cluster positions over the full sky
ra = np.random.random(N) * 360 # from 0 to 360 deg
sindec = np.random.random(N) * 2 - 1
dec = np.arcsin(sindec) * 180 / np.pi # from -90 to 90 deg
len(cluster_m)
Nstack
30
Background galaxy catalog generation
For each cluster of the ensemble, we use mock_data to generate a
background galaxy catalog and store the results in a GalaxyCluster
object. Note that: - The cluster density profiles follow the NFW
parametrisation - The source redshifts follow the Chang et
al. distribution and have associated pdfs - The shapes include shape
noise and shape measurement errors - Background galaxy catalogs are
independent, even if the clusters are close (i.e., no common galaxy
between two catalogs). - For each cluster we then compute - the
tangential and cross \(\Delta\Sigma\) for each background galaxy -
the weights w_ls to be used to compute the corresponding radial
profiles (see demo_compute_deltasigma_weights.ipynb notebook for
details)
The cluster objects are then stored in gclist.
import warnings
warnings.simplefilter(
"ignore"
) # just to prevent warning print out when looping over the cluster ensemble below
%%time
gclist = []
# number of galaxies in each cluster field (alternatively, can use the galaxy density instead)
n_gals = 10000
# ngal_density = 10
cosmo_clmm = Cosmology(H0=71.0, Omega_dm0=0.265 - 0.0448, Omega_b0=0.0448, Omega_k0=0.0)
# cosmo_clmm.set_be_cosmo(cosmo)
gal_args = dict(
cosmo=cosmo_clmm,
zsrc="chang13",
delta_so=200,
massdef="critical",
halo_profile_model="nfw",
zsrc_max=3.0,
field_size=10.0,
shapenoise=0.04,
photoz_sigma_unscaled=0.02,
ngals=n_gals,
mean_e_err=0.1,
)
for i in range(Nstack):
# generate background galaxy catalog for cluster i
cl = clmm.GalaxyCluster(
f"mock_cluster_{i:04}",
ra[i],
dec[i],
cluster_z[i],
galcat=mock.generate_galaxy_catalog(
cluster_m[i],
cluster_z[i],
conc_list[i],
cluster_ra=ra[i],
cluster_dec=dec[i],
zsrc_min=cluster_z[i] + 0.1,
**gal_args,
),
)
# compute DeltaSigma for each background galaxy
cl.compute_tangential_and_cross_components(
shape_component1="e1",
shape_component2="e2",
tan_component="DS_t",
cross_component="DS_x",
cosmo=cosmo_clmm,
is_deltasigma=True,
use_pdz=True,
)
# compute the weights to be used to bluid the DeltaSigma radial profiles
cl.compute_galaxy_weights(
use_pdz=True,
use_shape_noise=True,
shape_component1="e1",
shape_component2="e2",
use_shape_error=True,
shape_component1_err="e_err",
shape_component2_err="e_err",
weight_name="w_ls",
cosmo=cosmo_clmm,
is_deltasigma=True,
add=True,
)
# append the cluster in the list
gclist.append(cl)
CPU times: user 1.68 s, sys: 280 ms, total: 1.96 s
Wall time: 1.96 s
cl.galcat.columns
<TableColumns names=('ra','dec','e1','e2','e_err','z','ztrue','pzpdf','id','sigma_c_eff','theta','DS_t','DS_x','w_ls')>
Creating ClusterEnsemble object and estimation of individual excess surface density profiles
From the galaxy cluster object list gclist, we instantiate a cluster
ensemble object clusterensemble. This instantiation step uses - the
individual galaxy input \(\Delta\Sigma_+\) and
\(\Delta\Sigma_{\times}\) values (computed in the previous step,
DS_{t,x}) - the corresponding individual input weights w_ls
(computed in the previous step)
to compute
the output tangential
DS_tand cross signalDS_xbinned profiles (where the binning is controlled bybins)the associated profile weights
W_l(that will be used to compute the stacked profile)
%%time
ensemble_id = 1
bins = np.logspace(np.log10(0.3), np.log10(5), 10)
clusterensemble = ClusterEnsemble(
ensemble_id,
gclist,
tan_component_in="DS_t",
cross_component_in="DS_x",
tan_component_out="DS_t",
cross_component_out="DS_x",
weights_in="w_ls",
weights_out="W_l",
bins=bins,
bin_units="Mpc",
cosmo=cosmo_clmm,
)
CPU times: user 280 ms, sys: 0 ns, total: 280 ms
Wall time: 280 ms
There is also the option to create an ClusterEnsemble object without
the clusters list. Then, the user may compute the individual profile for
each wanted cluster and compute the radial profile once all the
indvidual profiles have been computed. This method may be reccomended if
there a large number of clusters to avoid excess of memory allocation,
since the user can generate each cluster separately, compute its
individual profile and then delete the cluster object.
ensemble_id2 = 2
empty_cluster_ensemble = ClusterEnsemble(ensemble_id2)
for cluster in gclist:
empty_cluster_ensemble.make_individual_radial_profile(
galaxycluster=cluster,
tan_component_in="DS_t",
cross_component_in="DS_x",
tan_component_out="DS_t",
cross_component_out="DS_x",
weights_in="w_ls",
weights_out="W_l",
bins=bins,
bin_units="Mpc",
cosmo=cosmo_clmm,
)
A third option is where all clusters already have the profile computed,
and we just add those to the ClusterEnsemble object:
# add profiles to gclist
for cluster in gclist:
cluster.make_radial_profile(
tan_component_in="DS_t",
cross_component_in="DS_x",
tan_component_out="DS_t",
cross_component_out="DS_x",
weights_in="w_ls",
weights_out="W_l",
bins=bins,
bin_units="Mpc",
cosmo=cosmo_clmm,
)
ensemble_id3 = 3
empty_cluster_ensemble = ClusterEnsemble(ensemble_id3)
for cluster in gclist:
empty_cluster_ensemble.add_individual_radial_profile(
galaxycluster=cluster,
profile_table=cluster.profile,
tan_component="DS_t",
cross_component="DS_x",
weights="W_l",
)
Stacked profile of the cluster ensemble
The stacked radial profile of the ensemble is then obtained as
clusterensemble.make_stacked_radial_profile(tan_component="DS_t", cross_component="DS_x")
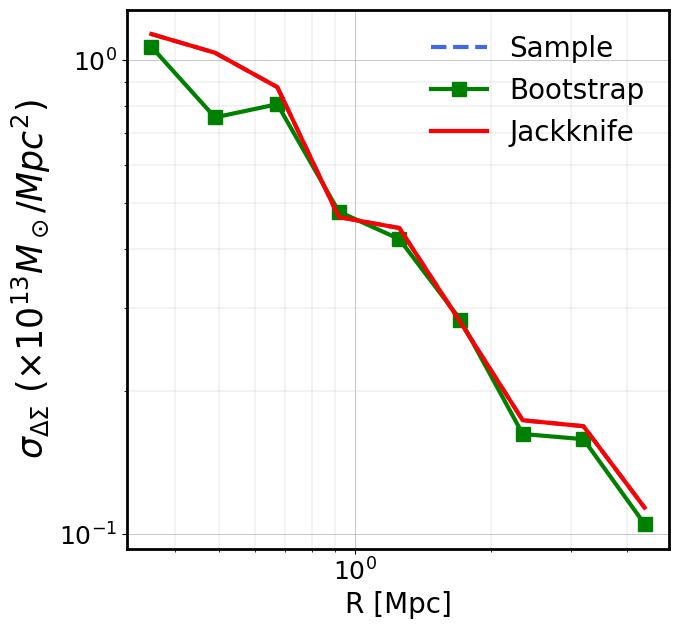
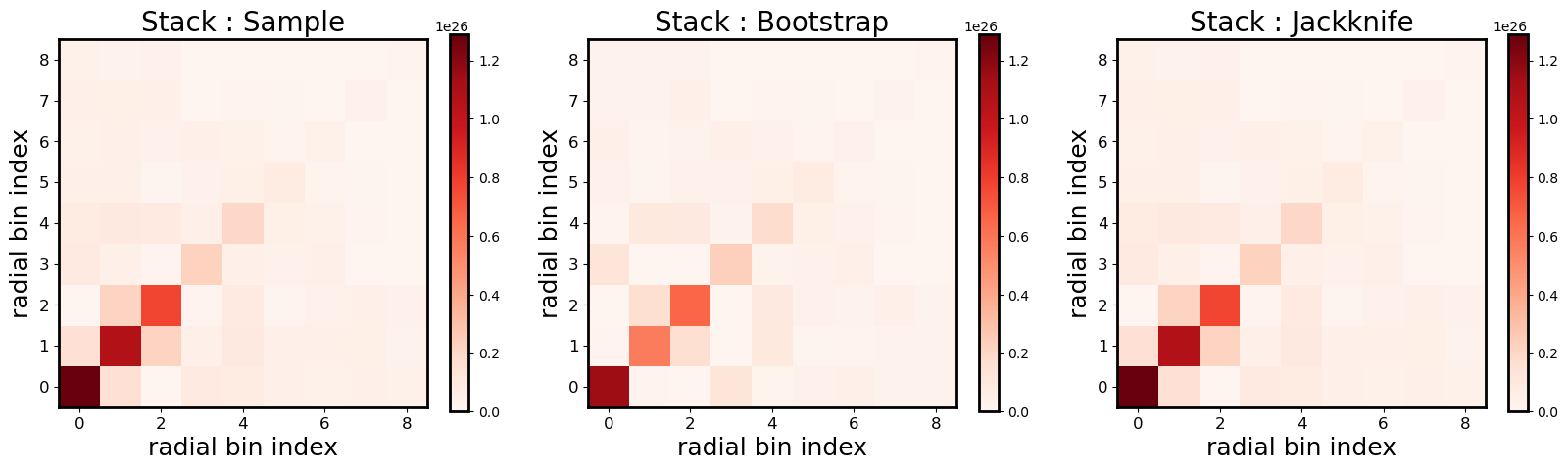
Covariance (Bootstrap, sample, Jackknife) of the stack between radial bins
Radial bins may be correlated and the ClusterEnsemble class provides
three methods to compute the covariance matrix of the stacked signal,
from the data: - The Sample covariance directly computes the covariance
between radial bins of the N individual cluster profiles of the
stack. - The Bootstrap approach is a resampling technique generating
n_bootstrap ensembles of N randomly drawn clusters from the
original ensemble, allowing for duplication. For each new ensemble, the
stacked profile is computed and the covariance computed over the
n_bootstrap stacks. - The Jackknife approach is another resampling
technique, that divides the sky in a given number of regions
\(N_{\rm region}\) and computes the covariance removing one region
(i.e the clusters of the ensemble in that region) at a time. The stack
is then computed using the remaining clusters and the covariance
computed over the \(N_{\rm region}\) number of stacks. The division
of the sky is done using the Healpix pixelisation scheme and is
controlled by the n_side parameter, with
\(N_{\rm region}=12 N_{\rm side}^2\).
NB: Approaches exist to compute the theoretical covariance of a stack
but these are not (yet) available in CLMM
clusterensemble.compute_sample_covariance(tan_component="DS_t", cross_component="DS_x")
clusterensemble.compute_bootstrap_covariance(
tan_component="DS_t", cross_component="DS_x", n_bootstrap=300
)
clusterensemble.compute_jackknife_covariance(
n_side=16, tan_component="DS_t", cross_component="DS_x"
)
plt.figure(figsize=(7, 7))
plt.rcParams["axes.linewidth"] = 2
plt.plot(
clusterensemble.data["radius"][0],
clusterensemble.cov["tan_sc"].diagonal() ** 0.5 / 1e13,
"--",
c="royalblue",
label="Sample",
linewidth=3,
)
plt.plot(
clusterensemble.data["radius"][0],
clusterensemble.cov["tan_jk"].diagonal() ** 0.5 / 1e13,
"-s",
c="g",
label="Bootstrap",
linewidth=3,
markersize=10,
)
plt.plot(
clusterensemble.data["radius"][0],
clusterensemble.cov["tan_sc"].diagonal() ** 0.5 / 1e13,
c="r",
label="Jackknife",
linewidth=3,
)
plt.xlabel("R [Mpc]", fontsize=20)
plt.ylabel(r"$\sigma_{\Delta\Sigma}\ (\times 10^{13} M_\odot /Mpc^2)$", fontsize=25)
plt.tick_params(axis="both", which="major", labelsize=18)
plt.legend(frameon=False, fontsize=20)
plt.loglog()
plt.grid(which="major", lw=0.5)
plt.grid(which="minor", lw=0.2)
fig, axes = plt.subplots(1, 3, figsize=(20, 5))
plt.rcParams["axes.linewidth"] = 2
fig.subplots_adjust(wspace=0.15, hspace=0)
maximum = clusterensemble.cov["tan_sc"].max()
for ax, cov, label in zip(
axes,
[clusterensemble.cov["tan_sc"], clusterensemble.cov["tan_jk"], clusterensemble.cov["tan_sc"]],
["Stack : Sample", "Stack : Bootstrap", "Stack : Jackknife"],
):
ax.set_title(label, fontsize=20)
ax.set_xlabel("radial bin index", fontsize=18)
ax.set_ylabel("radial bin index", fontsize=18)
ax.tick_params(axis="both", which="major", labelsize=12)
im = ax.imshow(cov, cmap="Reds", vmin=0, vmax=maximum, origin="lower")
plt.colorbar(im, ax=ax)
Visualizing the stacked profiles and corresponding model
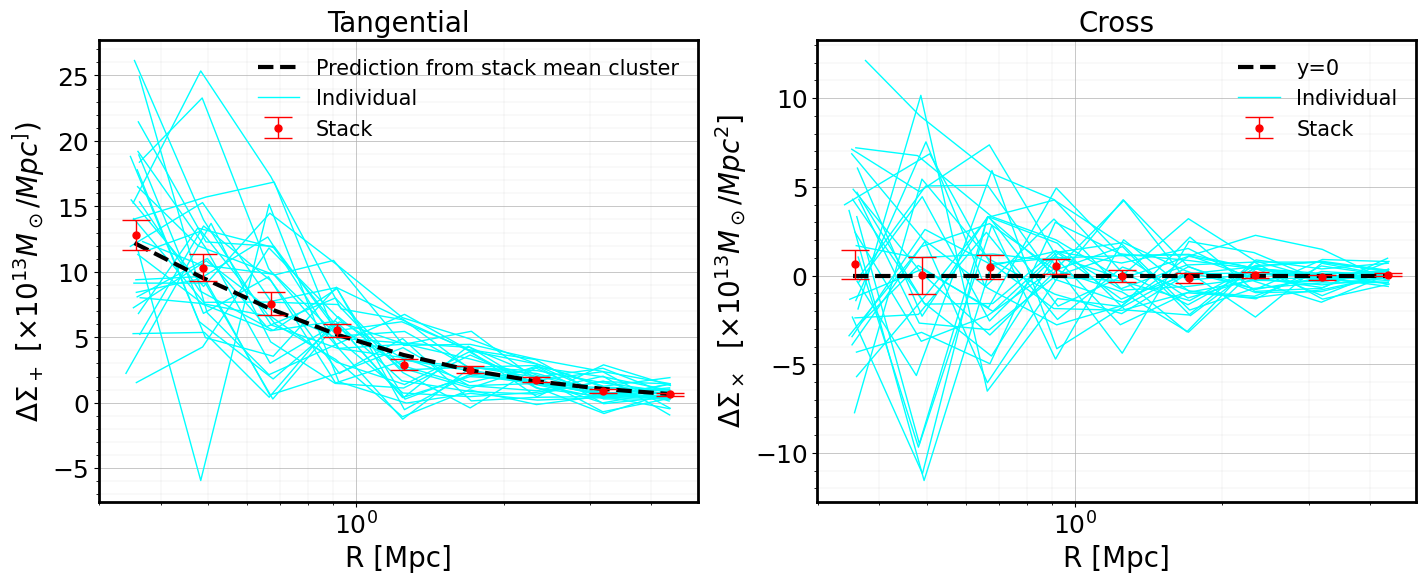
In the figure below, we plot: - the individual \(\Delta\Sigma\) profiles of the clusters (light blue) - the stacked signal (red symbols) - the prediction computed using a NFW profile and the mean values of the mass, concentration and redshift in the stack (dashed black)
moo = clmm.Modeling(massdef="critical", delta_mdef=200, halo_profile_model="nfw")
moo.set_cosmo(cosmo_clmm)
# Average values of mass and concentration of the ensemble to be used below
# to overplot the model on the stacked profile
moo.set_concentration(conc_list.mean())
moo.set_mass(cluster_m.mean())
r_stack, gt_stack, gx_stack = (clusterensemble.stacked_data[c] for c in ("radius", "DS_t", "DS_x"))
plt.rcParams["axes.linewidth"] = 2
fig, axs = plt.subplots(1, 2, figsize=(17, 6))
err_gt = clusterensemble.cov["tan_sc"].diagonal() ** 0.5 / 1e13
err_gx = clusterensemble.cov["cross_sc"].diagonal() ** 0.5 / 1e13
axs[0].errorbar(
r_stack,
gt_stack / 1e13,
err_gt,
markersize=5,
c="r",
fmt="o",
capsize=10,
elinewidth=1,
zorder=1000,
alpha=1,
label="Stack",
)
axs[1].errorbar(
r_stack,
gx_stack / 1e13,
err_gx,
markersize=5,
c="r",
fmt="o",
capsize=10,
elinewidth=1,
zorder=1000,
alpha=1,
label="Stack",
)
# print(moo.eval_excess_surface_density(clusterensemble.data['radius'][0], cluster_z.mean())/1e13)
axs[0].plot(
clusterensemble.data["radius"][0],
moo.eval_excess_surface_density(clusterensemble.data["radius"][0], cluster_z.mean()) / 1e13,
"--k",
linewidth=3,
label="Prediction from stack mean cluster",
zorder=100,
)
axs[1].plot(
clusterensemble.data["radius"][0],
0 * moo.eval_excess_surface_density(clusterensemble.data["radius"][0], cluster_z.mean()) / 1e13,
"--k",
linewidth=3,
label="y=0",
zorder=100,
)
axs[0].set_xscale("log")
axs[1].set_xscale("log")
for i in range(Nstack):
axs[0].plot(
clusterensemble.data["radius"][i],
clusterensemble.data["DS_t"][i] / 1e13,
color="cyan",
label="Individual",
alpha=1,
linewidth=1,
)
axs[1].plot(
clusterensemble.data["radius"][i],
clusterensemble.data["DS_x"][i] / 1e13,
color="cyan",
label="Individual",
alpha=1,
linewidth=1,
)
if i == 0:
axs[0].legend(frameon=False, fontsize=15)
axs[1].legend(frameon=False, fontsize=15)
# axs[0].plot(np.average(clusterensemble.data['radius'], axis = 0), np.average(clusterensemble.data['gt'], weights = None, axis = 0)/1e13)
axs[0].set_xlabel("R [Mpc]", fontsize=20)
axs[1].set_xlabel("R [Mpc]", fontsize=20)
axs[0].tick_params(axis="both", which="major", labelsize=18)
axs[1].tick_params(axis="both", which="major", labelsize=18)
axs[0].set_ylabel(r"$\Delta\Sigma_+$ $[\times 10^{13} M_\odot /Mpc^])$", fontsize=20)
axs[1].set_ylabel(r"$\Delta\Sigma_\times$ $[\times 10^{13} M_\odot /Mpc^2]$", fontsize=20)
axs[0].set_title(r"Tangential", fontsize=20)
axs[1].set_title(r"Cross", fontsize=20)
for ax in axs:
ax.minorticks_on()
ax.grid(lw=0.5)
ax.grid(which="minor", lw=0.1)
plt.show()
Saving/Loading ClusterEnsemble
The ClusterEnsemble object also have an option for saving/loading:
clusterensemble.save("ce.pkl")
clusterensemble2 = ClusterEnsemble.load("ce.pkl")